Benchmarking : Smil vs scikit-image
Morphological Image Libraries
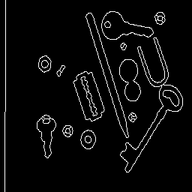
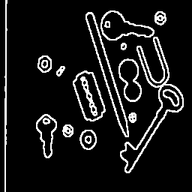
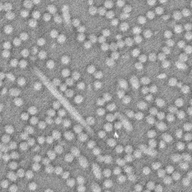
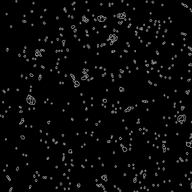
Watershed segmentationThis page shows how watershed segmentation is benchmarked. Both for binary and gray level images. The goal here isn't to obtain a perfect segmentation but to evaluate performance of the whatershed segmentation in a simple setup. As they're a little different, we compared a simple segmentation setup : as we do with Smil and as it's suggested at scikit-image documentation. See below. Qualitative results may not be exactly the same but are quite near good. As in the real life, some parameters may be fine tunned to give optimal results, we've just made a gross tunning, using the recommended methode we've found at scikit-image web site. For the presentation below, the images were post-processed in order to ease readability. Segmentation of gray level images - qualitative resultsSegmentation results
Segmentation methodBasically, both methods are similar : denoising, gross contour detection by gradient following a watershed. There were a little tunning on parameters h and sz for Smil and szg and sgo for scikit-image. The scikit-image result shows the basins instead of contours. In order to ease comparison with Smil, a post processing was done only for visualisation purposes in this page. The elapsed time to do this transformation wasn't take into account into results. Source code
Segmentation of binary imagesSegmentation of binary images is needed when one may want to semarate overlapping regions. From the test images, even if all images were benchmarked, only the two bellow meet this need. Segmentation results
Segmentation methodThe watershed algorithm may be used to segment only gray level images. To use it on binary images, the trick is to use some distance transform to create a gray level image from the binary one, invert it and use the max of the distance image as markers. For memory, the distance transform on a binary image replaces the value of each pixel by the distance to the nearest background pixel. While the algorithm proposed by scikit-image uses the Euclidean distance, Smil uses the morphological distance evaluated with an hexagonal structuring element. Source code
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||