Benchmarking : Smil vs scikit-image
Morphological Image Libraries
Benchmarking : what and how.
Presentation of resultsResults of the speedup benchmark are presented in three ways :
Results summary of elapsed times are shown here. What to evaluate and compareAs said in the introduction, we've choose to compare the indicators usually found in computational complexity comparison of algorithms : time and space. Elapsed Times (absolute elapsed times)Absolute elapsed times, by themselves, have no interest here as they strongly depend on many things, other than the algorithms, mainly on the platform they are evaluated. But the relative elapsed time may give interesting information about how application behavior change with the context (library, data size, data content, …) SpeedUp (relative elapsed times)The goal of this work is to evaluate how faster is Smil wrt scikit-image morphological functions. This can be defined as : 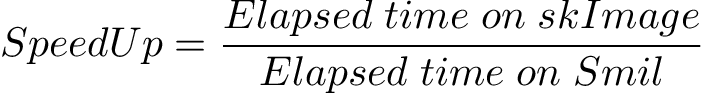
SpeedUp values greater than 1 mean Smil is faster than skImage. The opposite, if SpeedUp is smaller than 1. For some morphological operations, such as erode or opening, the elapsed time depends only on the image size. For others, such as segmentation or areaOpening, the elapsed time depends both on the image size and on its content. Surely, both kind of operations may also depend on the type and size of the structuring element, it there are one. Resources usage (space and CPU)It's important to have an idea of resources requirements. In one hand, they define how big or complex some specific hardware is able to hand and, in the another hand, one can evaluate how efficient (energy, …) the software is. In this version of benchmark we analized only two indicators : memory and CPU usage. See How resources usage is evaluated. This is the minimum we could do. Functions to benchmarkFunctions were chosen to cover most situations in the library :
The complete list of functions and their equivalent in scikit-image are enumerated in the function equivalence page. ImagesFunctions are benchmarked with 14 2D images of original size 256x256 or 512x512. For a list of all images, see Test Images). Test images were chosen in a way to be representative enough of usual applications. See more details at the images page. How elapsed time is evaluated
import timeit as tit
import math as m
import numpy as np
ctit = tit.Timer(lambda: sp.erode(imIn, imOut, se))
(nb, dt) = ctit.autorange()
if dt < 2.:
nb = m.ceil(nb * 2. / dt)
dt = ctit.repeat(repeat, nb)
# "dt" is a vector of length "repeat" with the
# cumulated time spent in each round.
elapsedMs = 1000 * np.array(dt).min() / nb
$ bin/smil-vs-skimage.py \
--function areaOpen --image hubble_EDF_gray.png \
--minImSize 256 --maxImSize 8192 \
--repeat 7 \
--selector min
How resources usage are evaluatedThe idea is similar to time elapsed evaluation but now a bigger image of size 16384x16384 image is created as a mosaic of 64x64 times the image lena.png. This means an image of size 256 MiB; Functions evaluated are open(), hMinima() and watershed(); Important : about the size of the mosaic image, an exception was done for function h_maxima() on the desktop computer (nestor), as the resident memory usage of skImage hit the installed memory size (16 GiB) and ran on swap. An extra run was done with a mosaic of size 32x32 (64 MiB). Both results are shown; Basically the procedure consists of two scripts run-mosaic.py which runs the function to be evaluated and pid-monitor.py which monitors, each second, cpu and memory usage by run-mosaic.py. This scripts are part of the repository smilBench; Typical usage of these scripts are as below :
$ bin/run-mosaic.py \
--function hMinima \
--imsize 16384 --ri 64 \
--repeat 5 \
--showpid --save \
--library smil \
images/lena.png
$ bin/pid-monitor.py --csv --pid 1431186 > usage-hMinima-lena-smil-16384.csv
About memory usage, it should be noticed that what's evaluated is the usage of the entire process. So this includes what's needed by python and all modules. Either way, evaluating Smil or skImage, the same python modules are loaded; We've done a simple experiment to evaluate the difference in memory usage when when the modules are loaded, or not, with no processing. The result is show in the discussion page. Other setup precisions
|