Benchmarking : Smil vs scikit-image
Morphological Image Libraries
DiscussionSummarySpeedUpGlobally, we can see from taurus and nestor summaries that Smil is always faster than scikit-image for gray level images. For binary images, scikit-image may win on functions doing labeling (label()) or depending on it. We can observe four situations about speedups in image sizes (see taurus and nestor summaries):
When varying the size of the Structuring Element, we observe that the speedup increases with the size. There are two reasons :
Both reasons have a big impact on the computational complexity as explained below at section Performance/Structuring Elements Resources usage (memory and CPU)We observed, with the help of an external application, the behaviour of python processes while running mathematical morphology operations. CPU usageAbout CPU usage, it's obvious that CPU usage is always intensive when handling huge data. The interest of what we saw was just to verify the effectiveness of parallelization in both libraries. While it was confirmed with Smil we haven't seen any sign of parallelization on Scikit-Image mathematical morphology operations. Looking around internet we haven't found any link about internal parallelization of Scikit-Image, but some hints on how to launch Python tasks. But this isn't related to parallelization at the library level and isn't transparent to the final user. But maybe we've lacking any information on how to enable parallelization at library level for Scikit-Image. Memory footprintWe observed that for both libraries the memory footprint of the Python process is much higher than the image memory size, even considering the eventual use of temporary allocated images. So, it's not related to libraries, but very probably to Python itself. The interesting information we've got with this experiment is that Scikit-Image always needs more memory than Smil. This imposes some limit on the image size each library can handle on computers with limited memory. We've reached a limit on the desktop computer (nestor), which has only 16GiB of installed memory for the function h_minima(). In this computer the image size limit for Scikit-Image was around 12000x12000 (144 MBytes), while the limit for Smil was around 32000x32000 (1 GBytes). Notes
Some programming issues impacting performanceLet's talk just about programming issues. Some strange, in appearence, behaviors can be seen on the graphics results, or even for small images. Most of the time these behaviors can be explained by hardware or operating system bottlenecks, such as memory bandwidth or cost, launching threads for small image sizes or synchronization issues in parallel code. ParallelismParallelism is implemented in Smil in two ways :
Combining both types of parallelism explains why Smil can be orders of magnitude faster than Scikit-Image, depending on the number of processor cores. Structuring elementsRepresentation and handling of structuring elementIn this work, all operations needing a structuring element, the default one was the choice : CrossSE() for Smil and diamond() for Scikit-Image, represented below when its size is 1 : 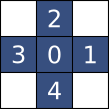
There are two differences between the way this structure is handled in the two libraries with significant impact in their performance :
If we report this to the complexity - number of memory accesses (r and w) - for each cell in the image (for 2D images and square bounding box for the structuring element), we have : 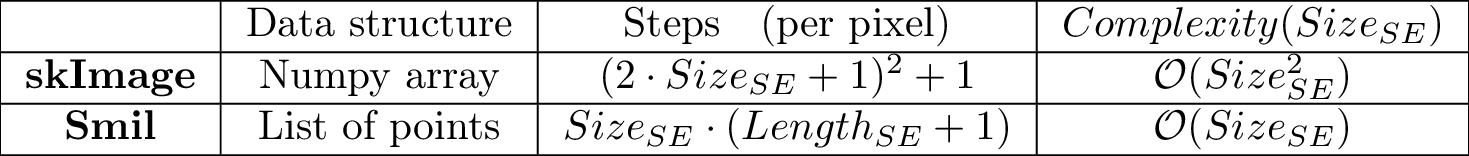
Structuring element versus connectivityWhile Smil always uses a formal structuring element on its morphological operations, Scikit-Image sometimes uses a connectivity parameter defined as: Maximum number of orthogonal hops to consider a pixel/voxel as a neighbor. Accepted values are ranging from 1 to input.ndim. Note : Equals to 1 for 4-connectivity and 2 for 8-connectivity on 2D Images. For 3D images, 3 can be used to full 26-connectivity, but 2 may cause some ambiguity in its interpretation. Source : scikit-image - Module: morphology Although this is convenient for 2D images and satisfies most situations, it doesn't allow for generic structuring elements nor for hexagonal grids. On the other hand, it allows the use of an optimized algorithm (See [XXX]) and that's why Scikit-Image label() function is faster than the Smil equivalent. Size of data typesAll tests here were done with images of type UINT8. A rule of thumb, which is valid really most of the time, is that “the bigger the data size the longer will be its processing time”. There are two main reasons :
To be convinced about the impact of the size of data types, try the following script in your computer :
import numpy as np
import timeit
d = 4096
print('{:27s} {:^7} {:^7} {:^7} {:^7}'.
format('', '+', '-', '*', '/'))
for t in [np.uint8, np.uint16, np.uint32, np.float32, np.float64]:
x = 70 * np.ones((d, d), dtype=t)
y = 3 * np.ones((d, d), dtype=t)
dp = 10 * timeit.timeit(lambda: x + y, number=100)
ds = 10 * timeit.timeit(lambda: x - y, number=100)
dm = 10 * timeit.timeit(lambda: x * y, number=100)
dd = 10 * timeit.timeit(lambda: x / y, number=100)
fmt = '* ({:d},{:d}) - {:8s} : '
fmt += '{:7.3f} {:7.3f} {:7.3f} {:7.3f} - ms'
print(fmt.format(d, d, str(np.dtype(t)), dp, ds, dm, dd))
You'll notice that, except for division, the elapsed time grows almost linearly with the data size. Division algorithm is, most of the time, iteractif and dependent on the values being handled. Next steps
|